Abstract: Implicit neural representations (INRs) have achieved remarkable successes in learning expressive yet compact signal representations. However, they are not naturally amenable to predictive tasks such as segmentation, where they must learn semantic structures over a distribution of signals. In this study, we introduce MetaSeg, a meta-learning framework to train INRs for medical image segmentation. MetaSeg uses an underlying INR that simultaneously predicts per pixel intensity values and class labels. It then uses a meta-learning procedure to find optimal initial parameters for this INR over a training dataset of images and segmentation maps, such that the INR can simply be fine-tuned to fit pixels of an unseen test image, and automatically decode its class labels. We evaluated MetaSeg on 2D and 3D brain MRI segmentation tasks and report Dice scores comparable to commonly used U-Net models, but with \(90\%\) fewer parameters. MetaSeg offers a fresh, scalable alternative to traditional resource-heavy architectures such as U-Nets and vision transformers for medical image segmentation.
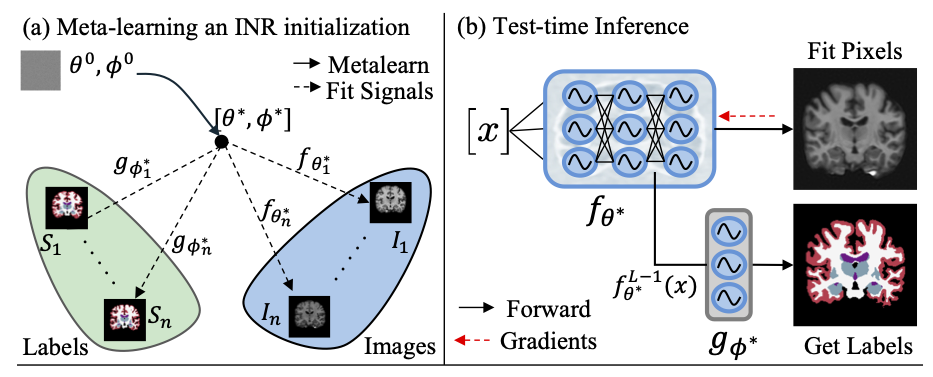
We use a meta-learning framework (shown in (a)) to learn optimal initial parameters \(\theta^*, \phi^*\) for an INR consisting of an \(L\)-layer reconstruction network \(f_\theta(\cdot)\) and shallow segmentation head \(g_\phi(\cdot)\). At test time (b), optimally initialized INR \(f_{\theta^*}\) is iteratively fit to the pixels of an unseen test scan. After convergence, the penultimate features \(f_{\theta^*}^{L-1}(x)\) are fed as input to the segmentation head \(g_{\phi^*}(\cdot)\) to predict per-pixel class labels.
MetaSeg Architecture
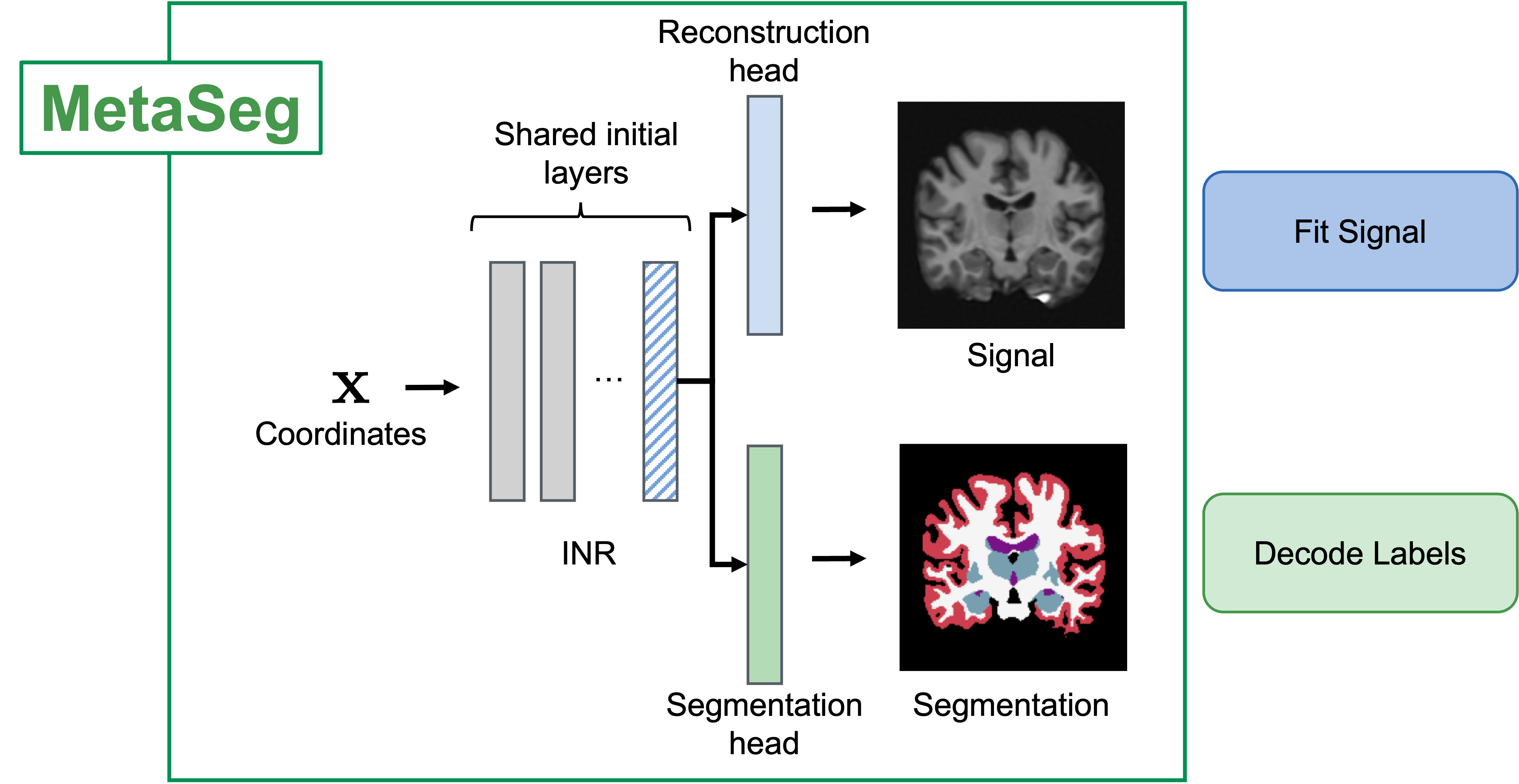
We use a Siren-INR where we share the initial layers of the INR are shared across a reconstruction head that fits the signal and a segmentation head which decodes learned INR features into per-pixel segmentation maps. We learn a semantically generalizable INR initialization for the INR and segmentation head using a meta-learning framework (Section 2 of paper)*. At test time, we fit the INR to the pixels of an unseen image and use the learned features to predict segmentation maps.
Test-time: Fit pixels, get labels!
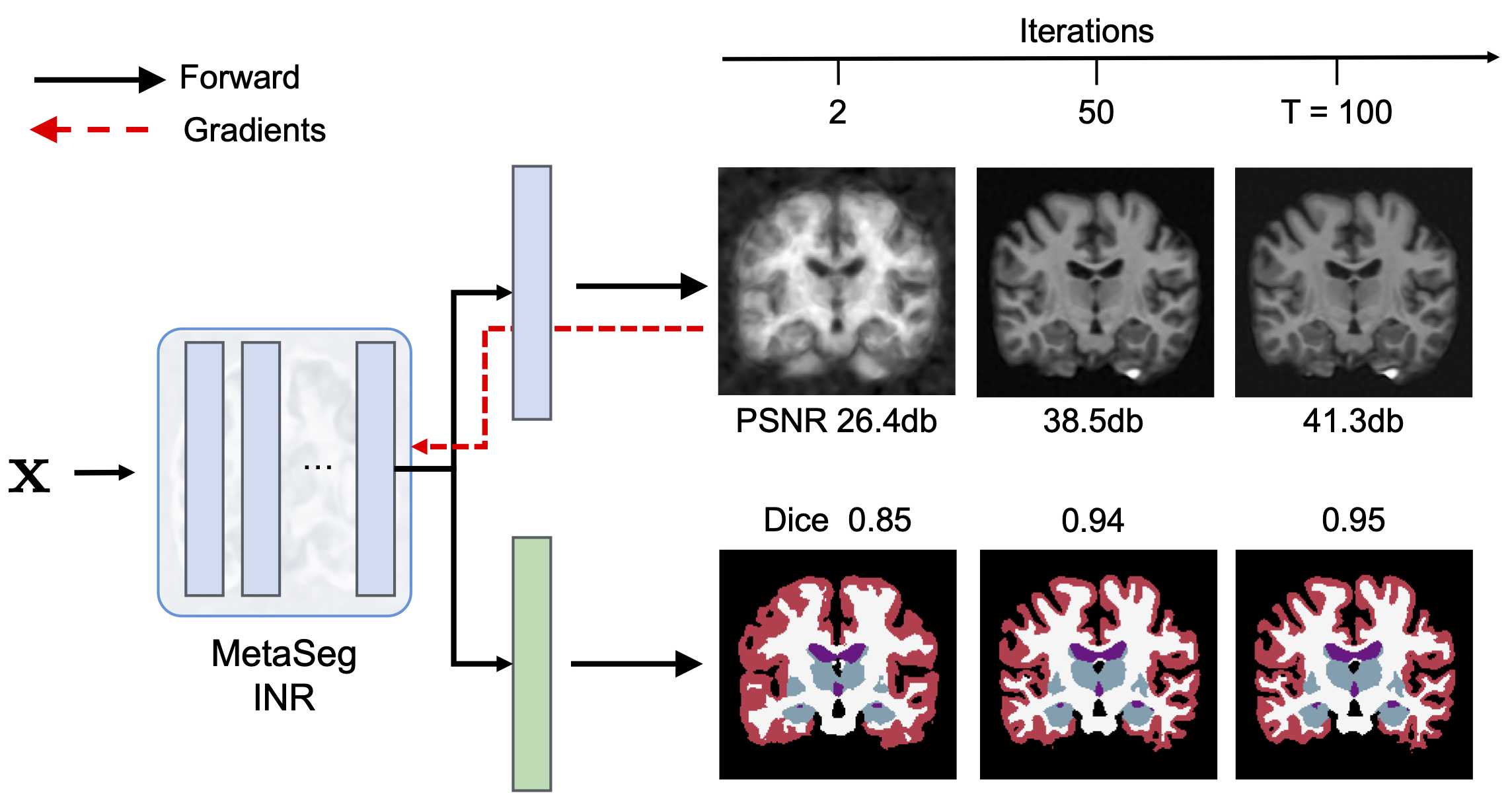
At test-time, MetaSeg initialized INRs along with its frozen segmentation head iteratively only fit the signal (intensity) while the segmentation head simultaneously decodes learned INR features of each pixel into a per-pixel segmentation map. it’s that simple: Fit pixels, to get labels! Further, thanks to the meta learned initalization, MetaSeg fits the signal faster and better than a randomly initialized INR (refer ablation table and discussion section in paper).
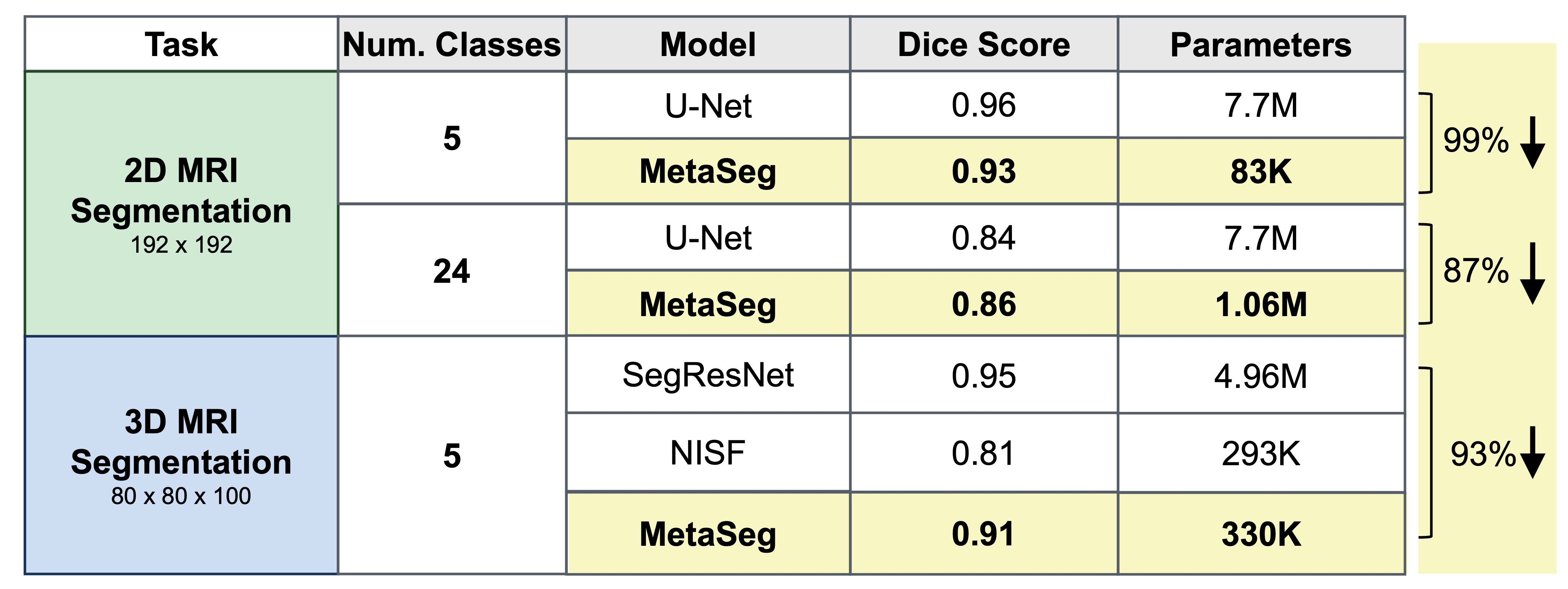
MetaSeg achieves comparable segmentation to U-Nets with 90% fewer parameters
Generalization of MetaSeg to small and large structures
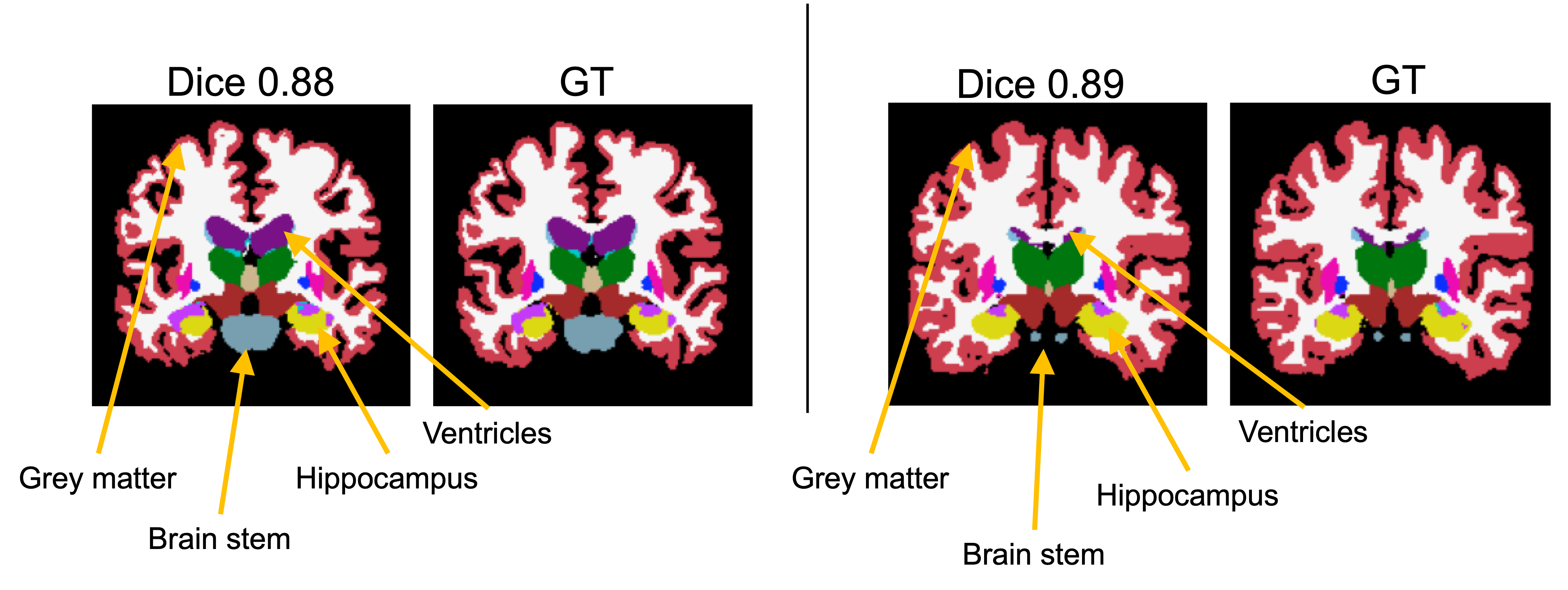
We further depict qualitatively, that MetaSeg’s semantic initialization generalizes well to varying brain anatomies, accurately accommodating large and small structures. Here we observe that high structural variances shown in the grey matter and brain stem are easily captured by MetaSeg. Deeper and delicate structures such as the hippocampus and ventricles (which can be a few pixels wide) are also segmented accurately by MetaSeg.
High quality 3D segmentation and generating cross-sectionv views with MetaSeg
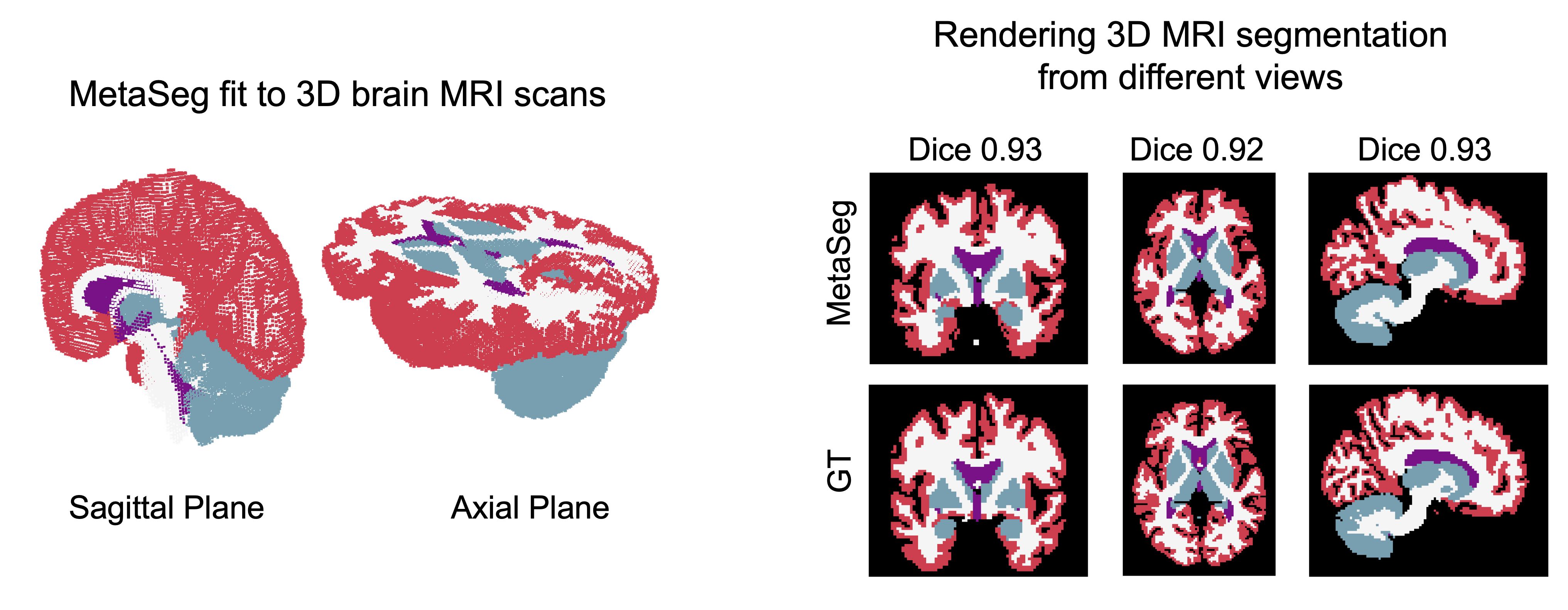
MetaSeg also scales to 3D signals readily and yields high quality 3d segmentation by only fitting 3D MRI scans. Further, since INRs are naturally interpolating models, we can query MetaSeg to obtain the segmentation of any section of the brain. As shown, MetaSeg can generate high quality cross sections with a dice of 0.93, which is in high agreement with the ground truth
Analysis of learned MetaSeg features
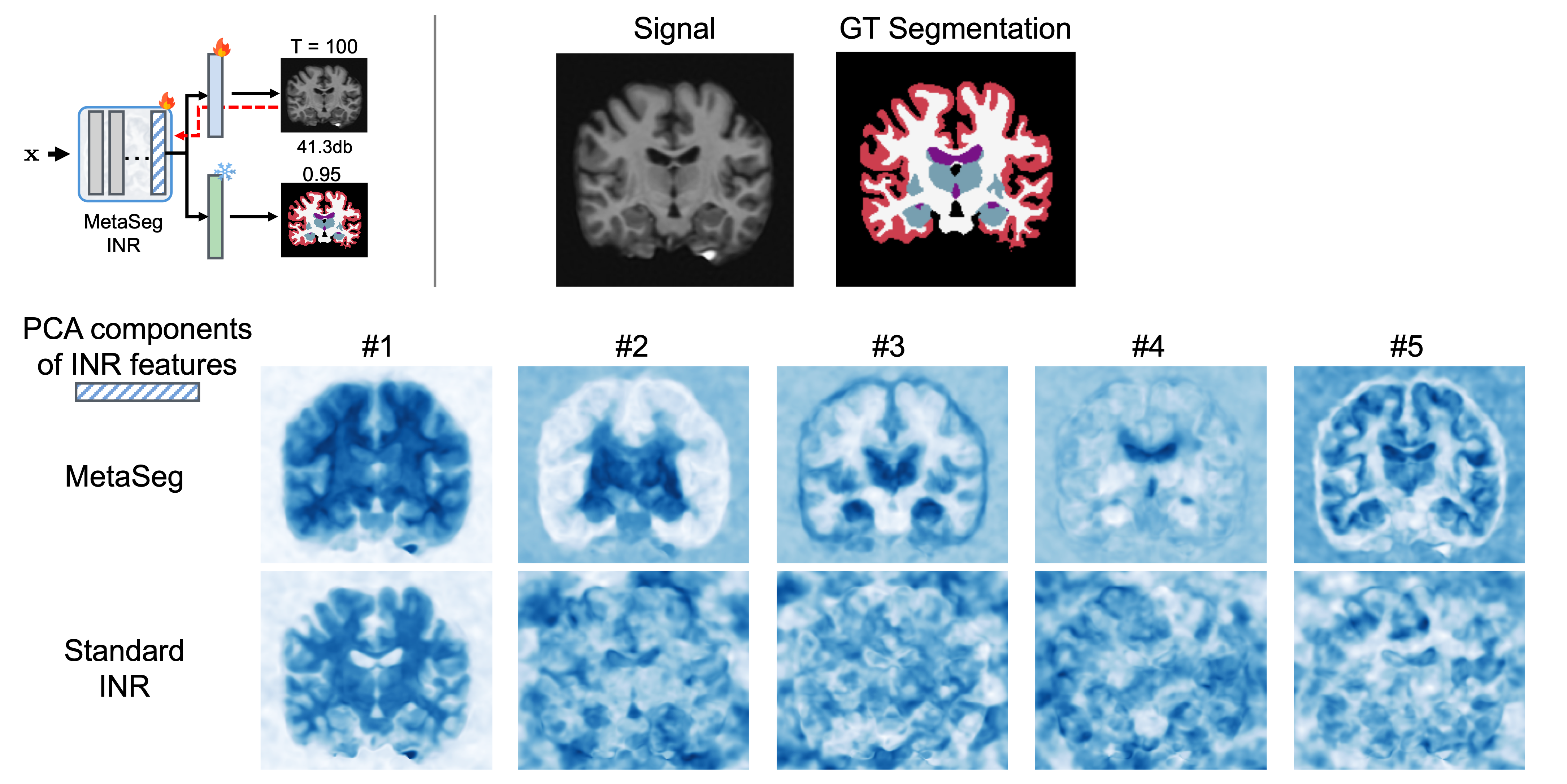
MetaSeg has shown impressive performance on 2D and 3D segmentation. To further understand the benefits of MetaSeg’s initialization and design, we visualize the principal components of MetaSeg’s learned INR feature space. We find that unlike a typical INR which would only fit to intensity, yielding seeming random deep principal components, MetaSeg’s feature space embeds semantic regions. As seen in components 3-5, regions such as ventricles, hippocampus, and white-grey matter boundary is strongly encoded in MetaSeg’s feature space.
For more details, please refer full paper!
Citation¶
@InProceedings{vyas2025metaseg,
author="Vyas, Kushal
and Veeraraghavan, Ashok
and Balakrishnan, Guha",
title="Fit Pixels, Get Labels: Meta-learned Implicit Networks for Image Segmentation",
booktitle="Medical Image Computing and Computer Assisted Intervention -- MICCAI 2025",
year="2026",
publisher="Springer Nature Switzerland",
pages="194--203",
isbn="978-3-032-04947-6"
}


